Recently, the research group led by Dr. Qiwei Qin from CAS key laboratory of marine bio-resources sustainable utilization (affiliated to South China Sea Institute of Oceanology, Chinese academy of sciences) made innovative progress in studying the marine virus encoded microRNAs. Team member Dr. Yang Yan et al identified and characterized a series of functional miRNAs encoded by a novel marine virus-Singapore grouper iridovirus (SGIV) for the first time. These findings have been recently published online in the world famous peer-reviewed journal PLoS ONE(Yan et al. 2011. Identification of a Novel Marine Fish Virus, Singapore Grouper Iridovirus-Encoded MicroRNAs Expressed in Grouper Cells by Solexa Sequencing. PLoS ONE. 2011. 6(4): e19148.)
MicroRNAs (miRNAs) are ubiquitous non-coding RNAs that regulate gene expression at the post-transcriptional level. An increasing number of studies has revealed that viruses can also encode miRNAs, which are proposed to be involved in viral replication and persistence, cell-mediated antiviral immune response, angiogenesis, and cell cycle regulation. Singapore grouper iridovirus (SGIV) is a pathogenic iridovirus that has severely affected grouper aquaculture in China and Southeast Asia. Comprehensive knowledge about the related miRNAs during SGIV infection is helpful for understanding the infection and the pathogenic mechanisms.
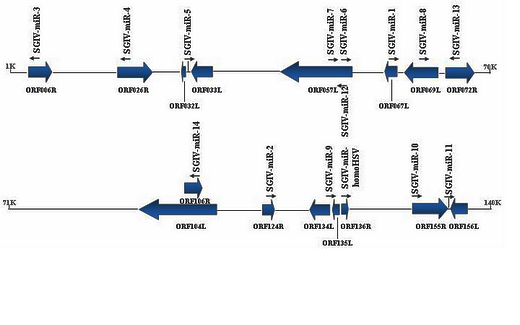
To determine whether SGIV encoded miRNAs during infection, a small RNA library derived from SGIV-infected grouper (GP) cells was constructed and sequenced by Illumina/Solexa deep-sequencing technology. They recovered 6,802,977 usable reads, of which 34,400 represented small RNA sequences encoded by SGIV. Sixteen novel SGIV-encoded miRNAs were identified by a computational pipeline, including a miRNA that shared a similar sequence to herpesvirus miRNA HSV2-miR-H4-5p, which suggests miRNAs are conserved in far related viruses. Generally, these 16 miRNAs are dispersed throughout the SGIV genome, whereas three are located within the ORF057L region. Some SGIV-encoded miRNAs showed marked sequence and length heterogeneity at their 3' and/or 5' end that could modulate their functions. Expression levels and potential biological activities of these viral miRNAs were examined by stem-loop quantitative RT-PCR and luciferase reporter assay, respectively, and 11 of these viral miRNAs were present and functional in SGIV-infected GP cells. The study provided a genome-wide view of miRNA production for iridoviruses and identified 16 novel viral miRNAs. To the best of our knowledge, this is the first experimental demonstration of miRNAs encoded byaquatic animal viruses. The results provide a useful resource for further in-depth studies on SGIV infection and iridovirus pathogenesis.
This work was supported in part by grants from the Natural Science Foundation of China (30725027, 30930070), the National Basic Research Program of China (973) (2006CB101802) and the Knowledge Innovation Program of Chinese Academy of Sciences (KZCX2-YW-BR-08, KZCX2-EW-Q213). Drs. Yang Yan and Huachun Cui are the first two authors of this PLoS ONE paper, and Prof. Qiwei Qin is the corresponding author.