Polycyclic tetramate macrolactams (PTMs) are a widely distributed family of natural products with diverse bioactivities. The structure complexity of PTMs is expanded by the fusion of a tetramate-containing macrolactam ring with a variety set of carbocyclic rings. Although a number of PTM biosynthetic gene clusters have been characterized or implicated in diverse bacteria, the biochemical mechanisms for the construction of polycyclic ring systems in PTMs remain enigmatic. Now, researchers at South China Sea Institute of Oceanology (SCSIO) reported the first biochemical mechanism for polycycle formation by a Micheal addition like reductive cyclization reaction, using ikarugamycin as a model (published in the issue of May 5, Angewandte Chemie International Edition, 2014, 53(19): 4840-4844).
Ikarugamycin is a PTM family member and has attracted extensive attentions as a promising lead in anticancer therapeutics since its discovery in early 1970s. Recently, researchers at SCSIO have reisolated ikarugamycin (1, Figure 1) from a marine Streptomyces sp. ZJ306 derived from the Pearl River estuary. Isotope labeling studies confirmed that ikarugamycin biosynthesis was originated from an unusual hybrid polyketide synthase/nonribosomal peptide synthetase (PKS/NRPS), consistent with other PTMs. Subsequently, dissection and reconstitution of 1 biosynthesis by in vivo gene disruption and heterologous expression studies not only identified a minimal three-gene cassette ikaABC capable of conferring the heterologous production of 1, but also established the function and reaction timing of IkaABC (Figure 1). Finally, the biochemical mechanism of the construction of an inner five-membered ring in 1, catalyzed by the NAD(P)H-dehydrogenase IkaC, was unveiled by elegant labelling studies using stereospecifically deuterated NADPH cofactor and/or deuterium oxide, which led to a mechanistic proposal involving in an unusual [1 + 6] Michael addition reaction (Figure 1).
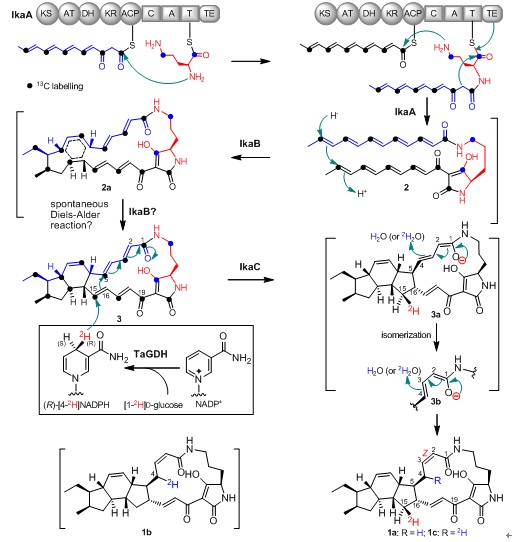
Figure 1. The proposed ikarugamycin (1) biosynthetic pathway and the IkaC catalytic mechanism. TaGDH, the glucose dehydrogenase from Thermoplasma acidophilum ATCC 25905.(Image by SCSIO)
This study provides the first biochemical evidence for reductive cyclization reactions in polycycle formation in PTMs and suggests that the same reductive cyclization strategy as IkaC catalysis may be generally applicable in the formation of polycyclic rings in other PTMs.
Drs. Guangtao Zhang and Wenjun Zhang are co-first authors of this study. This work is supported in part by the National Natural Science Foundation of China, the Ministry of Science and Technology of China, the Chinese Academy of Sciences and the Administration of Ocean and Fisheries of Guangdong Province.