Toxin/antitoxin (TA) systems are widespread in bacteria and archaea, and play prominent roles in cell physiology including altruistic behavior such as phage inhibition, global gene regulation, and tolerance to antibiotics. The cellular targets of these toxins are quite diverse and include DNA replication, mRNA stability, protein synthesis, cell-wall biosynthesis, and ATP synthesis. These toxins are co-expressed and neutralized with their cognate antitoxins from a TA operon in normally growing cells. Five different types of TA systems have been characterized, depending on the interaction of the TA and the nature of the antitoxin. Dr. WANG Xiaoxue has been working on TA systems for many years, and have identified and reported the first Type V TA system in E. coli [Nature Chemical Biology, 2012, 8(10): 855-861].
Dr. Wang’s group at the South China Sea Institute of Oceanology (SCSIO) continue to explore the physiological roles of TA systems in E. coli and in marine bacteria. For TA systems identified in E. coli so far, no toxin has been identified that functions by cleaving DNA. Dr. GUO Yunxue, a postdoctoral researcher from Dr. Wang, demonstrated via in intro studies that toxin RalR functions as a non-specific endonuclease that cleaves methylated and unmethylated DNA. Guo also demonstrated via mutagenesis that the antitoxin RNA is a trans-encoded small RNA (sRNA) with 16 nucleotides of complementarity to the toxin mRNA, and further determined the size of RalA by primer extension followed by qRT-PCR verification. Though RNA-protein EMSA and toxicity assays, Guo showed that the RNA chaperone Hfq was shown to be required for RalA antitoxin activity and appears to stabilize RalA. Moreover, collaborated with researchers at Pennsylvania State University, a beneficial role RalR/RalA to E. coli has been discovered with protection for host when treated with antibiotic fosfomycin. These results provide further evidence that the genes of cryptic prophage impact cell physiology and that these genes may be used to increase resistance to stress.
The unconventional type I TA systems identified by Guo et al. are more limited in distribution than typical type I TA systems, and are found mainly in Escherichia, Shigella, and Salmonella species. By comparing the phylogenetic tree of those identified type I loci with the host taxonomy, earlier studies suggest that type I loci have not been freely disseminated by horizontal gene transfer but instead may have a common ancient ancestor. Since TA genes from bacterial prophages are actively expressed in the human gastrointestinal tract, TA systems may play a dynamic role in more ecosystems than we expected. Further work on studying TA systems in marine bacteria will help to elucidate the roles of toxins and antitoxins in different marine ecosystems.
This study was published by Nucleic Acids Research [2014, 42 (10): 6448-6462], and this work is supported by National Basic Research Program of China [2013CB955701]; National Natural Science Foundation of China [NFSC31270214, NFSC31290233]; National Institutes of Health [NIH R01 GM089999];1000-Youth Elite Program (the Recruitment Program of Global Experts in China.
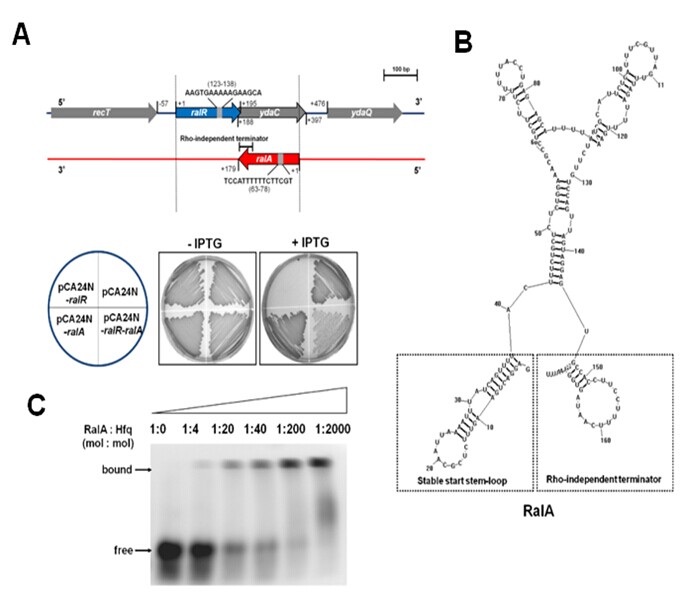
(A) RalR cleaves methylated DNA and unmethylated lambda DNA (dam-, dcm-). (B) RNA structure of RalA predicted by RNASTRUCTURE 4.6, with a stable start stem-loop at 5’ end and a Rho-independent terminator at 3’ end. (C) EMSA show that Hfq binds to RalA sRNA and the binding of Hfq to RalA increases with increasing Hfq.(Image by SCSIO)