The deep sea was assumed as a biological desert, because of extreme variations in pressure, salinity, and temperature. The recent advancements in marine technologies have allowed human-beings to access the deep sea and have led to the detection of microbial activities even in the Marina Trench with a depth of 10000 m. More and more culturable actinomycetes have been isolated and identified from deep-sea-derived sediment samples. Genome sequencing has revealed that most deep sea actinomycetes encode multiple biosynthetic gene clusters with little being connected to defined chemical structures. Genome mining, transcriptome mining, proteomics and a combination of these techniques have successfully activated some silent gene clusters into new metabolites. However, the traditional methods of changing culture conditions are still a powerful strategy for discovering new natural products. During the past ten years, researchers in South China Sea Institute of Oceanology(SCSIO), Chinese Academy of Sciences, have collected numerous culturable actinomycetes from deep sea samples obtained from open voyages to the South China Sea and Indian Ocean supported by National Natural Science Foundation of China. Researchers have systematically examined the metabolite profiles of a deep-sea-derived Streptomyces sp. SCSIO 03032 and have isolated and identified a total of 21 natural products including 15 novel ones.
Streptomyces sp. SCSIO 03032 was isolated by Dr. TIAN Xingpeng from a sediment sample at a depth of 3412 m from the Bay of Bengal in the Indian Ocean. When the strain was cultivated in the modified A1BFe + C medium, it was capable of producing four new bisindole alkaloids, spiroindimicins A-D, along with two known compounds lynamicins A and D. Spiroindimicins A-D possesses unusual spiro-ring systems and spiroindimicin B exhibited moderate cytotoxic activities against several tumor cell lines (Organic Letters,2012, 14, 3364-3367). Due to their unique structures, spiroindimicins A-D were selected in “Hot Off the Press” in Natural Product Reports (2012, 29, 1033-1037).
Upon cultivation in the same modified A1BFe + C medium, researchers found another two known a-pyridone antibiotics, piericidin A1 and mer-A2026B. Piericidin A1 was a potent inhibitor of both mitochondrial and bacterial NADH-ubiquinone oxidoreductase. By genome sequencing and bioinformatics analysis, researchers have identified the biosynthetic gene cluster for piericidin A1 and have elucidated tailoring biosynthetic steps including the PieE-catalyzed hydroxylation and PieB2-catalyzed methylation. Furthermore, a novel analogue piericidin E1 featuring a C2/C3 epoxy ring was identified from the pieE-disrupted mutant (Organic Letters 2014, 16, 734-739).
Most recently, 7 more bisindole alkaloids were isolated as trace components from Streptomyces sp. SCSIO 03032 cultivated in the modified A1BFe + C medium, including indimicins A-E, lynamicins F and G. Indimicins A–E bear a unique 1′, 3′-dimethyl-2′-hydroindole moiety. The C-methylation at the quaternary carbon C-3′, leading to the dearomatization of the indolocarbazole scaffold, or the C-methylation at C-2 of the pyrrole moiety in indimicins are rarely found in the L-tryptophan-derived bisindole alkaloids (Journal of Natural Products, 2014, DOI: 10.1021/np500362p).
To search for more compounds, researchers have investigated metabolite profiles of Streptomyces sp. SCSIO 03032 under cultivation in seven media, and they found three new compounds produced in modified ISP3 medium. These new compounds were characterized to be new macrolactams heronamides D-F (Journal of Natural Products 2014, 77, 388-391).
Streptomyces sp. SCSIO 03032 is a deep-sea-derived actinomycete strain and has been shown to potentially encode 26 biosynthetic gene clusters for secondary metabolites by genome sequencing. Researchers have been able to identify the gene clusters for synthesizing spiroindimicins and indimicins. Their detailed biosynthetic studies are still in progress to elucidate the unique enzymatic mechanism involved in the spiro-ring formation. In summary, these new structures from strain SCSIO 03032 not only expand the chemical variety of indole alkaloids, but also provide opportunities to investigate their biosynthetic origins at genetic and biochemical levels. Again, this work has revealed the worth of enhancing investigations on deep-sea-derived actinomycetes, for their great potential in discovering diversified and structure-unique novel natural products.
These studies were performed mainly by Drs. ZHANG Wenjun, LI Sumei, CHEN Yaolong, and ZHU Yiguang, and were financially supported by grants from National Natural Science Foundation of China, Ministry of Science and Technology of China, Chinese Academy of Sciences and Guangdong Province.
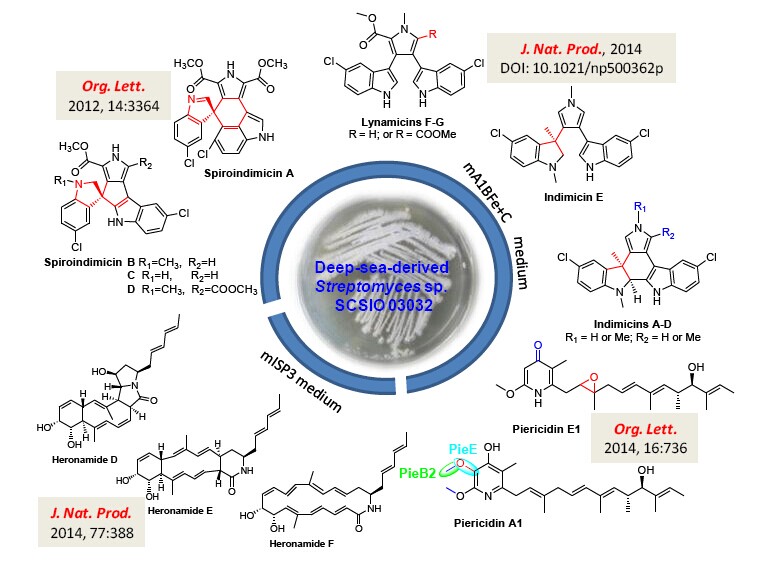
Figure 1. Novel metabolites produced by the deep-sea-derived Streptomyces sp. SCSIO 03032.(IMAGE BY SCSIO)
References
(1) Zhang W, Ma L, Li S, Liu Z, Chen Y, Zhang H, Zhang G, Zhang Q, Tian X, Yuan C, Zhang S, Zhang WM, Zhang C*.Indimicins A–E, Bisindole Alkaloids from the Deep Sea-Derived Streptomyces sp. SCSIO 03032. J. Nat. Prod., 2014, DOI: 10.1021/np500362p. [link]
(2) Zhang W, Li S, Zhu Y, Chen YC, Chen YL, Zhang HB, Zhang G, Tian X, Pan Y, Zhang S, Zhang WM, Zhang C*.Heronamides D–F, PolyketideMacrolactams from the Deep-Sea Derived Streptomyces sp. SCSIO 03032. J. Nat. Prod., 2014, 77(2): 388–391. [link]
(3) Chen Y, Zhang W, Zhu Y*, Zhang Q, Tian X, Zhang S, Zhang C*. Elucidating Hydroxylation and Methylation Steps Tailoring Piericidin A1 Biosynthesis. Org. Lett., 2014, 16(3): 736-739. [link]
(4) Zhang W, Liu Z, Li S, Yang T, Zhang Q, Ma L, Tian X, Zhang H, Huang C, Zhang S, Ju J, Shen Y, Zhang C*.Spiroindimicins A–D: New Bisindole Alkaloids from a Deep-Sea Derived Actinomycete. Org. Lett., 2012, 14(13): 3364-3367. [link]